Projects
Sympore - Plasmodesmata as Symplasmic Pores for Plant Cell-to-Cell Communication
During evolution of multicellularity, cells differentiated to become specialized and interdependent. Multicellular organisms invented channels for nutrient exchange and communication between cells. Plants uniquely developed plasmodesmata, complex cell-cell connections traversing the cell wall. Roles ascribed to plasmodesmata include selective transport of signals, ions, metabolites, RNAs and proteins. Due to technical hurdles, composition, structure and regulation of plasmodesmatal conductance remain enigmatic. Genetic approaches to study plasmodesmata were hampered by lethality or redundancy. Novel technologies now set the stage for resolving roles of plasmodesmata in transport and signaling in an interdisciplinary approach. We will use proximity labeling proteomics to obtain plasmodesmatal composition, and PAINT and cryo-electron tomography (cryoET) for near atomic structures. Models of plasmodesmata will be built from bottom up and top down approaches and combined with quantitative assessment of plasmodesmatal activity. Novel biosensor approaches together with knock down by genome editing will permit quantitation of transport of the diverse cargo. Single cell sequencing helps fine-tuning mutant selection and targeting of subtypes. Four labs will join forces: highly recognized experts in biophysics and cryoET (WB), advanced imaging and developmental signaling (RS), high-end proteomics and lipidomics (WS), and interactomics, transporters and cutting-edge biosensor technology (WF).
We will iteratively address: (1) systematic quantitative identification of components, (2) their localization and
dynamics, (3) structures and molecular building blocks of diverse plasmodesmatal types, and (4) transport and
signaling mechanisms. We expect breakthrough discoveries and completely new understanding of
plasmodesmatal function and evolution. Since plasmodesmata play key roles in nutrient allocation and virus
spread, we lay the basis for novel biotech solutions in agriculture.

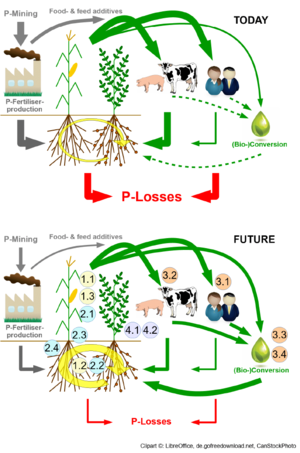
AMAIZE-P: Adaptation of maize-based food-feed-energy systesm to limited phosphate resources

RS 1.3 Regulatory modules of carbon resource allocation under different phosphate availabilities
Allocation of assimilated carbon to different tissues or organs is strongly influenced by nutrient availability. Especially phosphorus is a poorly available nutrient to plants due to fixation and extremely low diffusion rates of phosphates in the soil. Phosphate limitation generally enhances carbon allocation towards the root to produce a large root system. In addition, phosphate limitation stimulates the secretion of chelating agents such as citric acid and other carboxylates, or
the release of carbon to arbuscular mycorrhizal fungi in exchange for phosphate.
Thus, the uptake of phosphate from soils depends on sufficient carbon supply from the shoot to the root as well as on the carbon demand of other sink organs, such as the maize ear.
Potential topics for doctoral theses
Topic 1 (German): Establishing regulatory networks from large scale quantitative proteomic profiling in response to low phosphate availability
Topic 2 (German): Molecular basis of P use efficiency revealed from a comparative analysis of maize varieties with different P tolerance and carbon allocation properties
Topic 3 (German): From lab to field: Systematic validation of regulatory target genes in field experiments
Topic 1 (Chinese): Accumulation and allocation of starch in source and sink organs in maize under different P regimes
Topic 2 (Chinese): Spatiotemporal accumulation of carbohydrates and regulation of carbon metabolism in the maize ear under different P regimes
Topic 3 (Chinese): Regulation of carbohydrate transport by ZmPHR in the maize ear under P limitation
The Arabidopsis Kinome: Kinase-target relationships and proteome-wide analysis of protein phosphorylation sites
Plant Phosphorlyation Site Database PhosPhAt
Reversible phosphorylation is a key mechanism for regulating protein function. Thus it is of high interest to know which kinase can phosphorylate which proteins. Comprehensive information about phosphorylation sites in Arabidopsis proteins is hosted within the PhosPhAt database (http://phosphat.mpimp-golm.mpg.de). However, our knowledge of the kinases that phosphorylate those sites is dispersed throughout the literature and very difficult to access, particularly for investigators seeking to interpret large scale and high throughput experiments. Therefore, we aimed to compile information on kinase-substrate interactions and kinase-specific regulatory information and make this available via a new functionality embedded in PhosPhAt.
Our approach involved systematic surveying of the literature for regulatory information on the members of the major kinase families in Arabidopsis thaliana, such as CDPKs, MPK(KK)s, AGC kinases and SnRKs, as well as individual kinases from other families. To date, we have researched more than 4450 kinase related publications, which collectively contain information on about 289 kinases. Users can now query the PhosPhAt database not only for experimental and predicted phosphorylation sites of individual proteins, but also for known substrates for a given kinase or kinase family.
Nutrient induced phosphorylation dynamics and network
Most signalling pathways involve protein phosphorylation. Thus, modification-dependent changes in protein abundance and activity can be important features in signaling networks. Therefore, the analysis of phosphorylation events of membrane proteins under different nutrient regimes, and their dynamics was analyzed with the aim to identify proteins involved in early nutrient (carbon / nitrogen) signaling processes. Time courses of protein phosphorylation upon nutrient resupply after starvation was analyzed by quantitative protein mass spectrometry over a time course between 3 and 30 minutes.
In total, 1500 phosphorylation sites were identified under different nutrient conditions. Proteins with early phosphorylation changes (after 3 minutes and 5 minutes) involved GPI-anchored proteins, receptor kinases and transcription factors and other signaling proteins, while late responses (after 10 minutes and 30 minutes) involved protein synthesis and degradation as well as proteins with functions in metabolism. Our work provides novel candidate proteins with potential roles in nutrient perception and signaling.
Plasma membrane microdomain composition and their role in signaling processes
Membrane microdomains are based on local phase separation of membrane lipids and protiens. Particularly sterol- and sphingolipid-rich regions can biochemically be separated. We are interested in the functions of these membrane microdomains during membrane-induced signaling processes. Therefore, we used mutants disturbed at different stages in sterol biosynthesis pathways for proteomic analysis of membrane microdomains. As an analytical measure we observe alterations in protein abundance between detergent resistant and detergent soluble membranes, as well as between soluble proteins and intracellular membranes. We developed a systems approach including robust statistics involving bootstrapping to validate sterol-composition dependent membrane proteins and contaminant filtering using label free quantitation. The smt1 mutant showed a significant removal of the Signaling Proteins, Transporters and ATPases whereas mainly contaminants and co-purifying proteins were abundant in the smt1 DRM fraction. The ugt80A2;B1 mutant, which almost completely lacks glycosylated sterols, shows significant decrease of G-proteins and Aquaporins.
Quantitative proteomics method development
Since we are mainly interested in the protein functions, we rely on state-of the art quantitative proteomics. This field is still in development and given the diversity of biological questions to be addressed, we also need to develop new tools and methods in proteomics.
In the past we have made major contributions of setting up 15N-metabolic labeling as a precise quantitation method in plants. Current developments were also done with regards to data processing (cRacker).
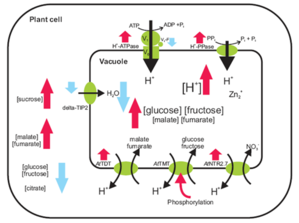
(finished) FOR1061: The role of tonoplast protein phosphorylation in activity regulation and protein targeting
The tonoplast membrane is an important compartment across which solute transport is tightly controlled under various environmental conditions. The goal of the proposed experiments is to elucidate the role of tonoplast protein phosphorylation in regulation of vacuolar solute accumulation and in protein targeting by the following experiments: (i) A targeted analysis by single reaction monitoring using specific labelled standard peptides of already known and predicted phosphopeptides and respective non-phosphopeptides will allow us to compare phosphorylation status and protein abundance. Up to 20 key transporters and channels across a wide range of stress conditions will be analyzed. (ii) An untargeted “discovery” approach using 15N-labeing will be carried out to identify proteins for which specific stress conditions or mutations of protein targeting pathways result in altered subcellular membrane location. Finally, different phosphorylation sites of the TMT protein family will be used as an example for (iii) in-depth characterization of the role of protein phosphorylation in stress-induced solute accumulation and protein targeting.